Question:
I have a problem with de novo assembly of my sequence reads. The problem manifests itself in different ways:
- Often, the assembly fails with following error message: "The following error occurred in the operator 'De novo assembly (Velvet)' (type 'De novo assembly (Velvet)'): Execution of the command "C:\[PATH here]\velvetg_mt" "C:\[PATH here]\action_88" -exp_cov auto -max_coverage 10000000 -cov_cutoff auto -min_contig_lgth 300' has been aborted (exit code 255)."
- In other occasions, the de novo assembly just seems to take forever.
- Sometimes the assembly actually completes, but with unsatisfactory results (many small contigs generated, few reads mapped).
Answer:
Could you please check the k-mer size that the Velvet de novo assembly algorithm is using? The default value might be too low for your read set. Reasonable values for k-mer length are 60-70% of the average read length, but make sure to enter an odd value.
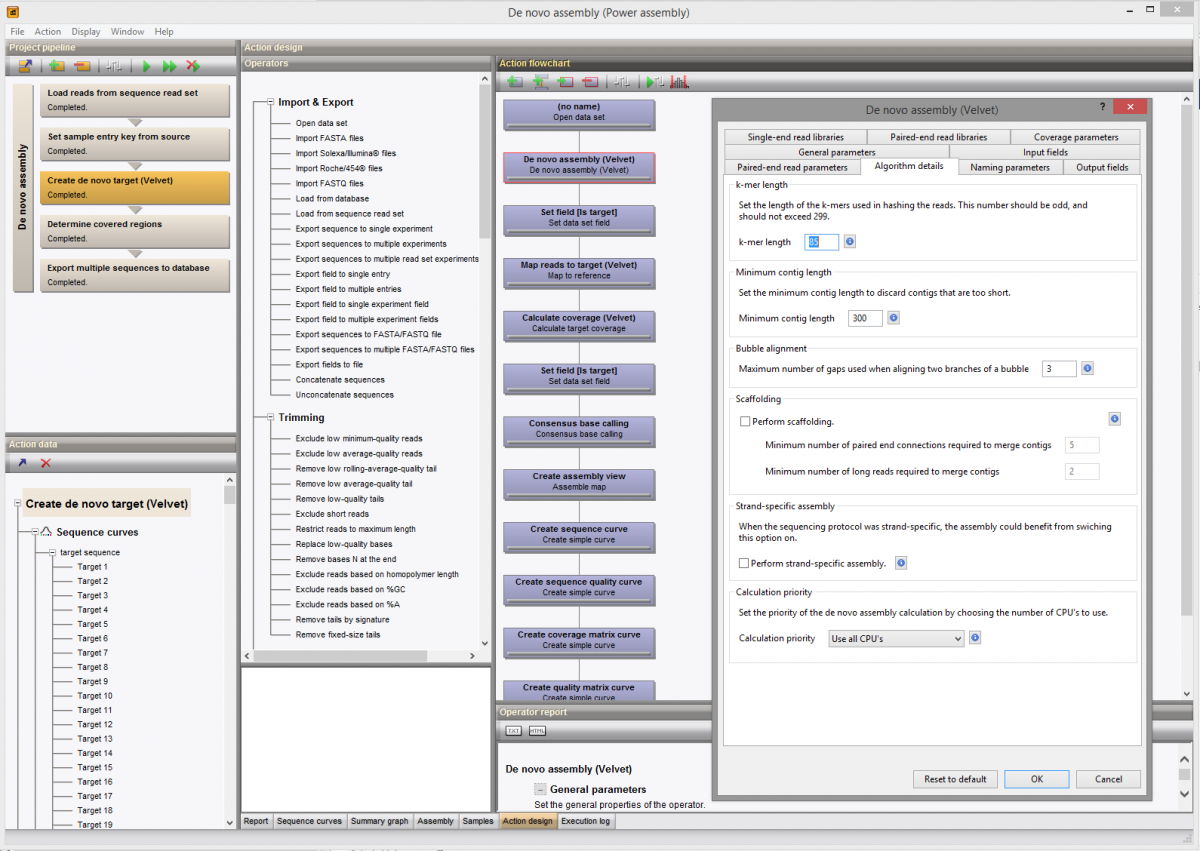
This is how to check the k-mer size in BioNumerics version 7.0 – 7.5:
- In the Power Assembly window, highlight the action Create de novo target (Velvet).
- Click on the Action design tab to activate the corresponding panel.
- Double-click the operator De novo assembly (Velvet) (second one in this action).
- In the dialog box that appears, click on the Algorithm details tab.
- The k-mer length is the first parameter in this tab.
From BioNumerics version 7.6 on, the k-mer size appears as a parameter in the wizard that appears when the de novo assembly is ran (runtime dialogs).
Package(s):
BIONUMERICS
Applicable for:
Version 7.0 - 8.1
